Edge-Directed Interpolation
Anyone who has done anything with graphics and/or image processing has had
to deal with image interpolation in one way or another. Most of the
graphics coders out there have dealt not far beyond the the bilinear texture
interpolation that is built into our 3d accelerator hardware. Even
so, most of us are able to recognize its weaknesses and cases where it produces
quite troublesome visual cues. Theoretically, we get some improvements
in more recent video cards by virtue of anisotropic filtering. However,
anisotropic filters serve more as a "don't make it look so bad" solution
rather than an "it will look good" solution. The interpolation itself
is faulty.
Well, the idea behind Edge-directed interpolation(EDI) is to use statistical
sampling to ensure quality of an image when you scale it up. This is
inasmuch as scaling is used on the actual image data to interpolate to a
new superresolution result as opposed to zooming in on an image, where you
actually WANT to see individual pixels. There were several earlier
methods that involved detecting edges to generate blending weights for linear
interpolation or classifying pixels according to their neighbour conditions
and using different otherwise isotropic interpolation schemes based on the
classification.
We've all seen the movies where FBI agents enhance an image so that faces
can be identified and so on... well, that's rather unrealistic, but
EDI is *something* like that. It's not going to produce features that
weren't there before, but what it DOES do is maintain features that originally
existed rather than making the image all blurry when it's scaled up.
The idea is to make sure that the image is NOT smooth perpendicular to edges
and it IS smooth parallel to edges. The first of the EDI algorithms
developed came from Jan Allebach and Ping Wong. While the Allebach/Wong
method does accomplish the main goals of EDI, it also tends to be overly
sensitive and produces noisy artifacts in the upsampled image. A newer
method came along from Xin Li and Michael Orchard which cured these problems
at a minor complication cost over linear interpolation. As I read the
original paper, though, it took a little deciphering on my part as far as
the equations... Not that they were complicated; They just suffered
from poor typesetting that made most of the math look bizzare. Ultimately,
though, it works out to a least-squares approximation.
So after a little mulling, I simplified the whole thing and worked out a
few optimizations as well as a handful of extensions to consider in the long
run.
The basic idea with Li's method is that edge directions are invariant to
resolution. If we were to actually solve for a curve that modelled
the edges of some image, we'd get basically the same curves for the same
image at any resolution. That is, within bounds of course, as some
small or short edges could be lost to extremely low resolution images.
However, actually solving for edge curves is a ridiculously slow process
involving PDE solvers... No fun. Moreover, it only tells us about
the image characteristics and not actual sampling information to draw from
for interpolation. However, statistical quantities, and in particular,
variances, will give us information about edges as high local variance quantities
means large changes in value -- i.e. an edge. Since we are dealing
with images (being that images are 2-d), we have two controllable variables
to to work with. Thus, the quantity in question becomes a covariance
character of pixel value with respect to x & y.
Any given interpolation approach boils down to weighted averages of neighbouring
pixels. The goal here is to make sure that we find optimal weights.
In the case of bilinear interpolation, we set all the weights to be equal.
In higher order interpolation methods like bicubic or sinc interpolation,
we consider a larger number of neighbours than just the adjacent ones.
In NEDI (New Edge-Directed Interpolation), we compute local covariances in
the original image, and use them to adapt the interpolation at high resolution.
Let us first consider the case of scaling a grayscale image with no interpolation
at all (this means not even nearest-neighbour).

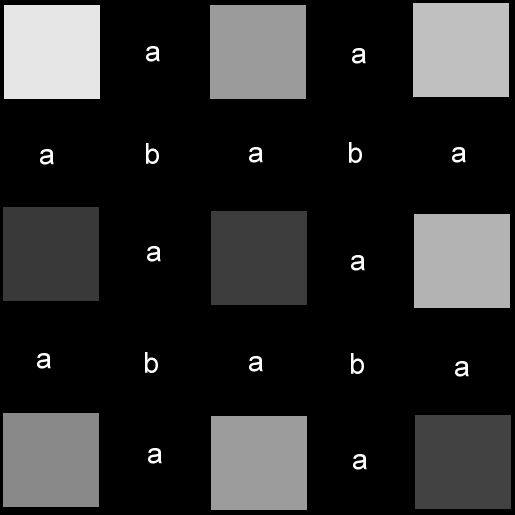
The image on the left represents a 3x3 image. Each square is a single
pixel and the spaces in between them are just for illustration. The
image on the right is scaled up to 6x6 (with the last column and row cropped
away to make it 5x5) with no interpolation. That is, we have 16 more
pixels in the superresolution image, but we've done nothing to give them
any values, so they simply remain zero-valued. As for the markings,
the pixels marked 'a' do not have all their adjacent neighbours
pre-determined. The pixels marked 'b' have pre-existing
neighbours... they just happen to be diagonal neighbours.
Fortunately, the question of diagonal or axial neighbours makes no difference
to NEDI. Covariances can be computed for any axis orientation, so for
the 'b' pixels, we simply use diagonal-axis covariances rather than xy-axis
covariances. The idea behind using covariance characters is that it
gives us a relative flow of differential characteristics by which to tune
the interpolator so that its support is along the edges only. NEDI
tends to preserve smoothness along edges of arbitrary orientation, and does
not produce smoothness perpendicular to the edges.
In practice, we obviously wouldn't concern ourselves with NEDI over a 3x3
image. Computing of covariances is done inside of a local window of
some size, MxM. Usually, we use at least a 4x4 window for reliable
estimation of covariances. The fundamental problem is simply taking
an HxW image and scaling it to a KHxKW image. For the sake of this
document, we'll stick to a case of K=2. For original image P, and
superresolution image Q, we will simply assert that P(x,y) = Q(2x, 2y).
This will at least solve for 25% of the pixels in the raster image of Q.
For the time being, we will stick to 4th order linear interpolation in our
final solution. That is, linear interpolation with only 4 neighbours
considered. The local covariances at the high resolution level can
be used to find the set of weights for optimal weighted linear interpolation
according to an MSE metric. Since we cannot directly have access to
the covariances at high resolution, we instead replace it with the low resolution
counterparts.
In theory, although we are sticking exclusively to the case of K=2, we can
actually achieve a scaling factor for all K=2k simply by repeating
the process k times. The only real problem with doing this is that
some accuracy is lost by the nature of using integer storage for images.
If scaling a float-HDR image, this is less of a concern because of the higher
precision. In an infinite-precision-float-based image format, repetition
would produce numerically identical results to special case implementations
for a particular value of K, assuming we run across no sensitivity
problems. If lack of precision errors are a concern, we can always
achieve a correct result by using a float image format in memory and only
rounding the final result. For cases like K=3 (that is, K not equal
to a power of two), we can achieve this simply two ways. We can use
NEDI for K=2, and bilinear interpolation to upsample for K = 1.5. The
other option is to use NEDI for K = 4, and then bilinear downsampling for
K = 0.75. Obviously, the former is faster, while the latter is higher
quality.
To perform our interpolation, we have to do so in two steps. First
we compute for the 'b' pixels, and then for the 'a' pixels. The 'b'
pixels have their 4 interpolation neighbours already known. The 'a'
pixels will only have 4 neighbours after 'b' pixels have been determined.
This was not a concern with isotropic interpolation schemes like bilinear
or bicubic because the location of a new pixel relative to the original pixmap
(the fractional uv coordinates) were all that was needed to determine the
appropriate weighting.
The 'b' pixels correspond to those that are at Q(2x+1, 2y+1) for the
superresolution image Q, where Q(2x, 2y) = P(x,y). By filling in the
values for 'b' pixels, we end up completing the lattice for all Q(i,j), where
(i+j) is even. This leaves only the 'a' pixels which are the set of
all Q(i,j), where (i+j) is odd.
To compute the ideal weights, we consider the following variables.
M -- local window of pixels is of size MxM
y -- A column vector of size M2 containing the pixels in the
window. y(k) is kth pixel in the window.
C -- A matrix of size 4xM2 containing the interpolation neighbours
of each pixel. The kth row vector contains the 4 interpolating neighbour
pixels of y(k).
1 1
R = --- * CT · C r = --- * CT · y
M*M M*M
R will end up being a 4x4 matrix, and r will be a column 4-vector.
Wiener filtering tells us that optimal MMSE linear interpolation weights
can be computed using the following.
a = R-1 · r = (CT · C)-1 * (CT · y)
The resulting 4-vector, a, will contain the
interpolating weights for the 4 neighbours. These weights are simply
multiplied by the corresponding neighbours and the results are summed together
to generate the new pixel. According to the paper by Li and Orchard,
the weights computed by using the diagonal neighbours of P(x,y) would be
the same weights used for Q(2x+1, 2y+1) ('b' pixel). Similarly, weights
computed using axial neighbours would also be used for the 'a' pixels.
The examples have been assuming a single channel so far, but the algorithm
can easily be extended to color images by simply using the same process for
all channels. For obvious reasons, the process cannot be used on indexed
color images for the same reasons that interpolation of any kind (excluding
nearest neighbour) cannot be used on indexed color images. Notably,
there are ways to improve performance on color images than to simply apply
the process once per channel on RGB color. For instance, using images
in colorspace that separates luminance and chrominance (e.g., YIQ, YUV, YCbCr)
means that we can limit EDI to only one channel. Namely, the luminance
channel Y. The chrominance channels can be handled with reasonable
quality by mere bilinear or bicubic interpolation (to be accurate, this holds
true for natural images, but not quite true for overtly graphical or 'cartoony'
images). RGB color would require the use of NEDI on all 3 channels
because each channel controls both luminance AND chrominance
simultaneously. Similarly, in HSL colorspace, we can afford bilinear
or bicubic on the H channel, but NEDI is warranted on the S and L channels.
Taking an example case here --

Let's say we're scaling up an image and what we have above is some small
section of the image. Now each intersection in the graph is a pixel,
and the dots are locations of new pixels in the superresolution image for
which we're trying to solve. These would be "b" pixels according to
our previous diagram. Now this 6x6 block actually corresponds to a
window size of M=4. Similarly, we'd actually need pixeldata from a
4x4 block for a window size M=2. Let's first consider a window size
of M=2, in which case the pertinent pixels are a 4x4 block in the upper left
corner. While I am numbering the pixels by their position in the subblock,
it should be noted that what we'd actually use in our computations are the
actual pixel values, be they RGB or grayscale or YUV or whatever. In
this case, we'll just assume that this is some section of a grayscale map
and the pixel numbers are also the actual grayscale levels of the pixels.
Let's take a superresolution pixel at the purple dot's location, and
use a window size of 2.
yT = [ 8 9 14 15 ]
[ 1 3 13 15 ]
[ 2 4 14 16 ]
C = [ 7 9 19 21 ]
[ 8 10 20 22 ]
The 2x2 window around the new pixel includes the pixels 8, 9, 14, and 15
(I use the transpose of y to save space because it's actually a column vector).
And since we're dealing with a "b" pixel, the interpolation neighbours
of consideration are the diagonal neighbours, so in the matrix C, each of
the row vectors are the 4 diagonal interpolation neighbours of each of pixels
in the 2x2 window. For instance, the diagonal neighbours of pixel 8
are pixels 1, 3, 13, and 15, and so on for pixels 9, 14, and 15.
For the case of a window size of 4, we'll need the entire 6x6 block, so I'll
build that example for the red dot in the diagram. The window will
include the 4*4 pixels surrounding the new superresolution pixel at the red
dot's location.
yT = [ 8 9 10 11 14 15 16 17 20 21 22 23 26 27 28 29 ]
[ 1 2 3 4 7 8 9 10 13 14 15 16 19 20 21 22 ]
[ 3 4 5 6 9 10 11 12 15 16 17 18 21 22 23 24 ]
CT = [ 13 14 15 16 19 20 21 22 25 26 27 28 31 32 33 34 ]
[ 15 16 17 18 21 22 23 24 27 28 29 30 33 34 35 36 ]
Note that I put the transpose of C here in this case to save some space.
So the aforementioned row vectors in the matrix C are column vectors
this time around. Again, we're dealing with "b" pixels, so we use the
diagonal neighbours of the pixels in the window. If we were working
on "a" pixels, we'd use the axial neighbours. For instance the axial
neighbours of pixel 8 would be pixels 2, 7, 9, and 14.
As you may have guessed, using a window of size 2 is a lot faster than using
a window of size 4. Also, more importantly, with larger windows, we
consider a larger neigbourhood and may lose some local characteristics --
in short, that makes for a more blurry result. This makes it sound
as if a smaller window is invariably better for NEDI. Unfortunately,
for us, with smaller windows comes more ill-conditioning of matrices, which
can lead to horrific errors in generating interpolation weights. Often
times, we see this in the form of stray pixels and weird misshapenness in
the resulting superresolution image. This happens a great deal with
regions that show severe gradients across edges.
As far as the original implementation of Li's NEDI algorithm is concerned,
this is all there is to it. However, there do exist further steps one
can take on top of the usual.
The simplest is a performance-oriented variation. Pre-computing weights
for all the pixels in the window of the original image. For M=4, we'd
have 16 pixels in an MxM window. We can pre-compute the 4-vectors of
weights for both axial neighbours and diagonal neighbours for all those 16
pixels and store them ahead of time. Even though neighbours of a pixel
in a window may be outside the window doesn't really matter because we will
only be considering those pixels for a given MxM block. By doing this,
we are only calculating weights for each pixel once.
Another variation is to interpolate weights. Every pixel in the original
image will generate its own set of weights based on its own local
covariance. This means that, for instance, direct replacement of the
weights for Q(2x+1, 2y+1) by the weights generated for Q(2x, 2y) may not
be absolutely correct. For every pixel, you are considering 4 interpolation
neighbours from which you take a weighted sum. In theory, we can consider
all 4 of those neighbours in the generation of weights. In short, we
take all the 4 interpolation neighbours and average the pre-computed set
of weights to generate a local weighting. The end result is that instead
of averaging pixels, as is done in bilinear interpolation, we are averaging
the covariance characters. In this case, you would act as if the current
image is a superresolution image based on a yet lower resolution image and
interpolate the blendweights.
Further still, we can increase the order of the linear interpolation.
As described above, the problem simply handles things by 4th order interpolation
because we're limited to scanning only 4 directions. The only neighbours
available to the high resolution image during generation are either
vertex(diagonal) neighbours or edge(axial) neighbours. However, we
can potentially increase the order of interpolation by including neighbours
that are farther down in those 4 directions. For example, even though
we're limited to scanning in 4 directions, we can check against 2 pixels
in each direction rather than one, thereby resulting in 8th order interpolation.
One of the nice advantages of using higher order interpolation is that
it naturally increases the size of the matrices involved and reduces chances
of ill-conditioning without having to use larger neighbourhoods. Of
course, it's a significantly higher computational cost, for instance, to
have to invert 8x8 matrices than 4x4.
And for the sake of example, I have some sample images taken straight from
Li's paper --

original
image |
Scaled up 16x (4x in each axis)

Bilinear interpolation |

Bicubic interpolation |

Allebach & Wong EDI |

Li & Orchard New EDI |







